Abstract
Introduction
Donor cell derived myeloid neoplasm (DCMN), defined as the development of de novohematological malignancies from cells of donor origin,is a late complication of allogeneic hematopoietic stem cell transplantation (allo-HSCT). We report on seven cases of DCMN in which whole-exome sequencing (WES) at different time-points after allo-HSCT, as well as in a sample from each donor, was performed. The ultimate objective was to accurately described the clonal architecture, spatial heterogeneity and identify somatic mutations that are induced in the process of leukemogenesis and clonal evolution of myeloid neoplasm. Donors were also analyzed to detect underlying condition predisposing to the development of DCMN.
Patient and Methods
Seven patients with a confirmed diagnosis of DCMN and their donors were recruited from different Spanish institutions. This cohort included a total of 32 BM samples at different time points after allo-HSCTand one PB sample from each donor (one case received dual allo-HSCT, CB and PB from both donors were obtained), which provided a total of 40 samples.
Genomic DNA samples were prepared according to Agilent SureSelect-XT Human exon 50Mb enrichment kit (Agilent Technlogies, Santa Clara, CA) preparation guide and libraries were sequenced on Illumina HiSeq platform (Illumina, San Diego, CA). DNA sequencing data from recipient post-transplant BM samples, were matched against their donor PB sample and previous post-transplant BM samples to identify the acquisition of mutations along the post allo-HSCT period.Germline variants in donors were studied in order to detect mutations that predisposed to the development of a myeloid neoplasm.The research protocol was approved by the Ethic Committee of Gregorio Marañón General University Hospital. Patients´ and donors´ information was collected from their medical records.
Results
Clinical and biological characteristics of the 7 patients with DCMN and their donors are shown in Table 1.
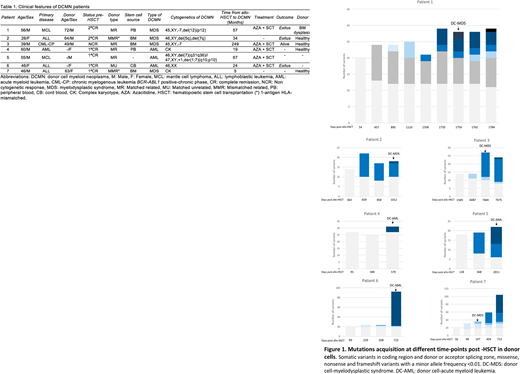
Mutational profiles obtained from the follow-up samples at different time-points post-HSCT demonstrated high intra-tumor genetic heterogeneity and clonal dynamic for all cases. The number of variants are increased over time and at the moment of DCMN diagnosis, the median number of variants was 28, ranging from 18 to 92 variants (Figure 1).
WES identified in DCMN patients gene mutations commonly seen in adult AML or MDS, such as in SETBP1, DNMT3A, TET2, RUNX1, CSF3R, EP300and IDH2.In addition, others non-silent variants were acquired in all cases. Among the additional novel alterated genes, we found 23 strong candidateswith oncogenic potential. LUC7L2, NOP14, LAMA5, SKOR2, EML1, SNX13, RHPN2, IRS1, MTG2, TENM2, MEFV, GSE1, NOTCH4, DTX1, CNOT4, PNKP, GRB7,SENP7,TAF1L, ZKSCAN2, ZBTB20, ZNF461 and MEGF10.
Analysis of CNVs revealed numerical alterations across the post allo-HSCT samples in patients 1, 2, 3, 4, 5 and 7. The most common chromosomal alterations in DCMN were monosomy 7 and other chromosome 7 abnormalities, which detected in the 86% (6/7) of the patients.
Although none of the donors developed a myeloid neoplasm at the moment of diagnosis of DCMN in recipient, donor 1 revealed an abnormal karyotype (45,XY,-7) at the moment of the allo-HSCT. All other donors harbored at least one pathogenic or probably-pathogenic variants, most probably of germline origin, in genes involved in hematological or solid tumor predisposition.
Conclusions
The development of DCMN involves dynamic genomic processes that begin months before the clinical onset. In this study, our integrated multi-step analysis revealed the intra-tumor heterogeneity and evolutionary history of seven DCMN.
The present study reveals a process of sequential clonal expansions promoted by the acquisition of somatic mutations in donor hematopoietic cells.
Detection of heritable or acquired gene mutations in donors associated with predisposition to haematological malignancies could have clinical implications for the patients undergoing to allo-HSCT.
Leukemic transformation ofdonor hematopoietic stem cells provides a useful in vivomodel to study the mechanisms involved in leukemogenesis.
Novel approaches based on high-depth next generation sequencing to study consecutive samples from post-transplant period in these patients, appear promising to discover new genes involved in myeloid neoplasm and to decipher the mechanisms of leukemogenesis.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal